Examples include RNAi-mediated formation of heterochromatin at non-coding and repeated DNA in Saccharomyces pombe, or transcriptional silencing promoted by the association of chromatin-remodeling complexes with ncRNA-binding proteins such as IDN2 in Arabidospsis thaliana or Nrd1 in S. pombe and S. cerevisiae. Whether a similar scenario also occurs in trypanosomes is not known at present. In this work we describe the characterization of the RNAbinding protein RBP33, which was first identified in a search for proteins that were able to bind to a uridine-rich RNA element in vitro. RBP33 depletion results in the accumulation of transcripts derived from silenced regions, such as SSRs and retroposon genes, and thus it could have a role in regulating gene silencing in trypanosomes. In this work, we have undertaken the characterization of the RNA-binding protein RBP33. We found 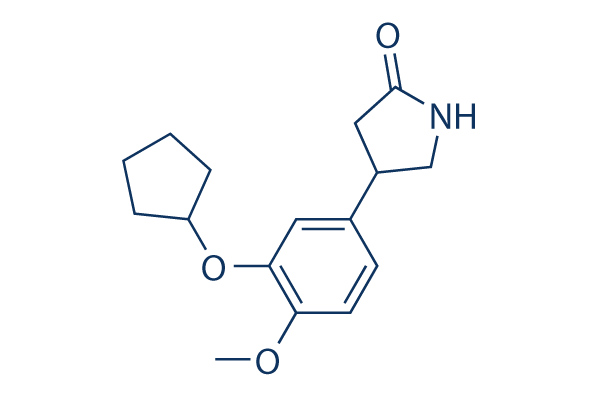 that the protein is localized in the nucleus, and is MDL-29951 essential in both bloodstream and procyclic trypanosomes. Using RNA immunoprecipitation and deep sequencing, we were able to identify a cohort of transcripts associated with RBP33 and thus obtain valuable information about the function of this protein. Although RBP33 binds to mRNAs encoding known or conserved proteins, most of its RNA targets are likely to be non-coding and/or present at minimal levels in the cell. Indeed, 90% of the RBP33-associated RNAs annotated as coding for ‘hypothetical proteins’ or ‘hypothetical proteins, unlikely’ were not detected by ribosome profiling, and 75% were not detected in a global transcriptome survey. Moreover, over one-third of the corresponding genes are located next to SSRs, close to the ends of chromosomes or have an antisense orientation within a transcription unit. Interestingly, depletion of RBP33 results in an accumulation of SSR-derived transcripts. In the related parasite Leishmania tarentolae, it has been shown that base J prevents read-through at RNA polymerase II termination sites. L. tarentolae cells seem to require base J for viability, but T. brucei can survive without J. Two thymidine hydrolases are Cinoxacin involved in the synthesis of J, but none is encoded by RBP33-bound mRNAs. To our knowledge, this is the first instance of a regulatory protein that has a role in silencing gene expression at RNA polymerase II termination regions. In addition, RBP33 silencing promotes the accumulation of retroposon and repeat-derived RNAs. This phenomenon has been also observed upon disruption of the RNAi machinery in T. brucei, raising the question of whether RBP33 could have a role in the regulation of the RNAi pathway in trypanosomes. In fact, convergent SSRs have been shown to produce small interfering RNAs in T. brucei. However, the molecular phenotype observed upon depletion of RBP33 presents some differences with that observed in trypanosomes lacking a functional RNAi pathway. Thus, deletion of the genes encoding the dicer-like protein TbDCL2 or the argonaute protein TbAGO1 results in the accumulation of full-length INGI transcripts which are,6 kb long and CIR147-derived transcripts of,10 kb, whereas the transcripts observed upon RBP33 silencing were smaller. We did, however, detect full-length transcripts corresponding to SLACs retroposons. Second, the accumulation of retroposon and repeat transcripts is stage-dependent, being almost undetected in procyclic trypanosomes. And third, deletion of TbDCL2 did not result in the accumulation of convergent SSRderived transcripts, whereas this sort of RNAs was readily visible upon RBP33 ablation.
that the protein is localized in the nucleus, and is MDL-29951 essential in both bloodstream and procyclic trypanosomes. Using RNA immunoprecipitation and deep sequencing, we were able to identify a cohort of transcripts associated with RBP33 and thus obtain valuable information about the function of this protein. Although RBP33 binds to mRNAs encoding known or conserved proteins, most of its RNA targets are likely to be non-coding and/or present at minimal levels in the cell. Indeed, 90% of the RBP33-associated RNAs annotated as coding for ‘hypothetical proteins’ or ‘hypothetical proteins, unlikely’ were not detected by ribosome profiling, and 75% were not detected in a global transcriptome survey. Moreover, over one-third of the corresponding genes are located next to SSRs, close to the ends of chromosomes or have an antisense orientation within a transcription unit. Interestingly, depletion of RBP33 results in an accumulation of SSR-derived transcripts. In the related parasite Leishmania tarentolae, it has been shown that base J prevents read-through at RNA polymerase II termination sites. L. tarentolae cells seem to require base J for viability, but T. brucei can survive without J. Two thymidine hydrolases are Cinoxacin involved in the synthesis of J, but none is encoded by RBP33-bound mRNAs. To our knowledge, this is the first instance of a regulatory protein that has a role in silencing gene expression at RNA polymerase II termination regions. In addition, RBP33 silencing promotes the accumulation of retroposon and repeat-derived RNAs. This phenomenon has been also observed upon disruption of the RNAi machinery in T. brucei, raising the question of whether RBP33 could have a role in the regulation of the RNAi pathway in trypanosomes. In fact, convergent SSRs have been shown to produce small interfering RNAs in T. brucei. However, the molecular phenotype observed upon depletion of RBP33 presents some differences with that observed in trypanosomes lacking a functional RNAi pathway. Thus, deletion of the genes encoding the dicer-like protein TbDCL2 or the argonaute protein TbAGO1 results in the accumulation of full-length INGI transcripts which are,6 kb long and CIR147-derived transcripts of,10 kb, whereas the transcripts observed upon RBP33 silencing were smaller. We did, however, detect full-length transcripts corresponding to SLACs retroposons. Second, the accumulation of retroposon and repeat transcripts is stage-dependent, being almost undetected in procyclic trypanosomes. And third, deletion of TbDCL2 did not result in the accumulation of convergent SSRderived transcripts, whereas this sort of RNAs was readily visible upon RBP33 ablation.