Both Rep1 and Rep2 required simultaneous mutation of multiple lysine residues to effectively block their sumoylation. Based on two-hybrid assays, the three sites Semaxanib mutated in Rep13R each likely represent sites targeted for SUMO conjugation; however, it seems unlikely that the thirteen residues mutated in Rep213R each normally serve as a SUMO acceptor site. Proteins targeted for RAD001 mTOR inhibitor sumoylation often contain multiple SUMO acceptor sites, but in other studies, abolishing major sumoylation sites has been shown to lead to increased SUMO conjugation at less preferred sites and this may also be the case for Rep1 and Rep2. Despite the multiple mutations in Rep13R and Rep213R that abolished two-hybrid interaction with SUMO, faint slower-migrating Rep protein species were detected in western blot analysis, suggesting sumoylation was still occurring, albeit at a low level, on remaining lysine residues. However, since inheritance of 2 mm plasmids encoding Rep13R or Rep213R mutants was perturbed, the residual sumoylation was not sufficient for normal Rep protein function. Sumoylation can have diverse effects, altering the activity, interactions, localization or stability of a targeted protein. Our analyses demonstrated that Rep1 and Rep2 were not dependent on sumoylation for their interaction with each other, consistent with observations that bacterially-expressed Rep1 and Rep2 interact in vitro. However, the Rep13R and Rep213R mutants were defective for association with the plasmid STB partitioning locus. Even mutation of a single sumoylation site in Rep1, K305, modestly reduced Rep1-STB association. Since efficient partitioning of the 2 mm plasmid has been shown to depend on association of Rep1 and Rep2 with STB, loss of association of Rep1 and Rep2 lysine-to-arginine mutants with STB is likely to be the primary defect leading to loss of efficient plasmid partitioning. Consistent with the hypothesis that impaired sumoylation of Rep1 correlates with defective assembly of the Rep1-Rep2 complex at STB, when Rep1 was sumoylation-deficient, both Rep proteins lost their localization to discrete nuclear foci previously shown to contain clusters of the 2 mm plasmid. The uniform nuclear staining pattern observed for sumoylation-deficient Rep1 is reminiscent of that observed for wild-type Rep1 in rsc2D yeast, in which association of Rep1 with STB is also impaired, supporting Rep1 interaction with STB as critical for Rep1 sub-nuclear localization. Sumoylation has been shown to regulate the sub-nuclear localization of other proteins, a notable example being the SUMO-dependent localization of promyelocytic leukemia protein to PML nuclear bodies in mammalian cells. In this study, 2 mm plasmid foci were observed  even when Rep1 and Rep2 were both sumoylationdeficient. Further investigation is needed to assess whether localization of plasmid clusters in their normal spindle poleproximal nuclear address is altered by changes in Rep1 and Rep2 sumoylation status. Our data suggest that Rep protein sumoylation may promote stable association of the Rep proteins with 2 mm plasmid DNA, an association reminiscent of sumoylation-dependent targeting of proteins to centromeres in yeast and in higher eukaryotes. In human cells, proteins conjugated with SUMO-2/3 are enriched at centromeres.
even when Rep1 and Rep2 were both sumoylationdeficient. Further investigation is needed to assess whether localization of plasmid clusters in their normal spindle poleproximal nuclear address is altered by changes in Rep1 and Rep2 sumoylation status. Our data suggest that Rep protein sumoylation may promote stable association of the Rep proteins with 2 mm plasmid DNA, an association reminiscent of sumoylation-dependent targeting of proteins to centromeres in yeast and in higher eukaryotes. In human cells, proteins conjugated with SUMO-2/3 are enriched at centromeres.
Author: small molecule
Both hosts contain enzymes of the biosynthesis pathway for ergosterol production from zymosterol
Both host trypanosomatids have glycerol kinase and 3-glycerophosphate acyltransferase, enzymes for the synthesis of 1,2-diacyl-sn-glycerol and triacylglycerol from D-glycerate. In endosymbionts, glycerolipid metabolism seems to be reduced to two enzymes: 3glycerophosphate acyltransferase and 1-acylglycerol-3-phosphate O-acyltransferase, suggesting metabolic complementation between partners. Furthermore, as well as the pathway of sterol biosynthesis that produces lanosterol from farnesyl-PP. These pathways are only complete in A. deanei. The symbionts do not have enzymes for sterol biosynthesis, in accordance with our previous biochemical analysis. Symbiosis in trypanosomatids is characterized by intensive metabolic exchanges, reducing the nutritional requirements of these trypanosomatids when compared to species without the Z-VAD-FMK Symbiotic bacterium, or to aposymbiotic strains. Several biochemical studies have been carried out analyzing the biosynthetic pathways involved in this intricate relationship as recently reviewed, and our genomic data corroborate these findings. A schematic description of the potential metabolic interactions concerning the metabolism of amino acids, vitamins, cofactors, and hemin is provided in Figure 7. Both symbiotic bacteria have genes potentially encoding for all necessary enzymes for lysine, phenylalanine, tryptophan and tyrosine synthesis, in agreement with previous experimental data. Tyrosine is required in the growth medium of A. deanei, but it is not essential for S. oncolpelti or S. culicis. Here, in the symbiotic bacteria, we found enzymes involved in tyrosine synthesis, as well as indications that phenylalanine and tyrosine can be interconverted. In fact, protozoan growth is very slow in absence of phenylalanine and tryptophan, which may indicate that larger amounts of these amino acids are required for rapid cell proliferation. Our data indicate that branched-chain amino acid synthesis mainly occurs in the symbionts except for the last step, with the branched-chain amino acid aminotransferase found in the host protozoan. Among the pathways that involve contributions from both partners, two have previously been characterized in detail, the urea cycle and heme synthesis. The urea cycle is complete in both symbiont-harboring trypanosomatids. Symbiotic bacteria SAR131675 msds contribute with ornithine carbamoyltransferase, which converts ornithine to citrulline, and with ornithine acetyltransferase, which transforms acetylornithine in ornithine. Conversely, aposymbiotic strains and symbiont-free Crithidia species need exogenous arginine or citrulline for cell proliferation. Our genomic data corroborate these studies. Contrary to symbiont-free trypanosomatids, A. deanei and S. culicis do not require any source of heme for growth because the bacterium contains the required enzymes to produce heme precursors that complete the heme synthesis pathway in the host cell. Our results support the idea that heme biosynthesis is mainly accomplished by the endosymbiont, with the last three steps of this pathway performed by the host trypanosomatid, and in most cases also by the bacterium as described in. Furthermore, this metabolic route may represent 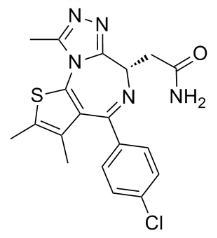 the result of extensive gene loss and multiple lateral gene transfer events in trypanosomatids.
the result of extensive gene loss and multiple lateral gene transfer events in trypanosomatids.
The photosynthesis involving the reduction of the carbon dioxide into sugars is a well-known redox process
Thus, elevated expressions of the transcripts belonging to the photosynthesis in “off” year in relation to the “on” year were observed for citrus. Besides, the photosynthesis was inhibited by the bud morphology in the “on” year, whereas “off” year leaves were filled with photoassimilates. In fact, it has been proposed that its induction in “off” citrus buds provides a leaf signal indicating the available nutrition richness. Similarly, the pistachio trees accumulated more carbohydrate during “off” years in relation to the “on” ones. Thus, Goldschmidt supported the regulatory role of the photoassimilate availability for the flowering induction. The comparison between the mature bearing and 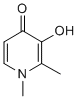 non-bearing leaves also showed that the ion transport- and homeostasis-related transcripts were more expressed in the non-bearing leaves than in the bearing ones. The KEGG analyses of those up-regulated transcript in the non-bearing leaves indicated that they were mainly involved in the lipid and amino acid metabolism, xenobiotic biodegradation and metabolism, biosynthesis of secondary metabolites, and carbohydrate and energy metabolism. Interestingly, the xenobiotic biodegradation and metabolism participates in the defense mechanisms. Indeed, a relationship between the carbohydrate nutritional status and the responses to the xenobiotics has been found in Arabidopsis thaliana, showing that the presence of sugars triggered the defense mechanisms. On the other hand, the flavonoids controlling the aroma and flavor are secondary metabolites, being synthesized in response to an excess of photoassimilation. The expression level of the transcripts related with flavonoid biosynthesis were increased in the “off” year leaves in relation to the “on” year ones in the olive tree. A similar gene expression pattern was observed with citrus buds, being based on flavonoids acting as a reservoir for the photoassimilation surplus. Taking account the higher expression of transcripts related to the oxidation-reduction, carbohydrate metabolism and mineral transport, together with flavonoid biosynthesis in “off” year leaves, we conclude that the nutritional status may be the principal key controlling the alternate bearing in the olive tree. Supporting the present work, a wide range of the genes targeted by the olive tree miRNA were found to be mainly involved in the carbohydrate metabolic pathways. Despite providing no selective advantage to the host, the plasmid is faithfully maintained at,60 copies per cell. The 2 mm plasmid achieves this by encoding copy-number amplification and partitioning systems, and by borrowing host cell machinery for its replication and segregation. Retention of the 2 mm plasmid at normal copy number is also dependent on the host cell process of sumoylation, the post-translational modification of target proteins with the small Orbifloxacin ubiquitin-related modifier protein SUMO. Sumoylation is an essential conserved eukaryotic function known to regulate diverse cellular processes by modulating the interactions, localization, or post-translational stability of substrate proteins. Like ubiquitin, SUMO must be activated in a series of enzyme-catalyzed steps Tulathromycin B before being conjugated to target proteins by the E2 conjugating enzyme Ubc9. Some target proteins require an E3 ligase for recruitment to Ubc9.
non-bearing leaves also showed that the ion transport- and homeostasis-related transcripts were more expressed in the non-bearing leaves than in the bearing ones. The KEGG analyses of those up-regulated transcript in the non-bearing leaves indicated that they were mainly involved in the lipid and amino acid metabolism, xenobiotic biodegradation and metabolism, biosynthesis of secondary metabolites, and carbohydrate and energy metabolism. Interestingly, the xenobiotic biodegradation and metabolism participates in the defense mechanisms. Indeed, a relationship between the carbohydrate nutritional status and the responses to the xenobiotics has been found in Arabidopsis thaliana, showing that the presence of sugars triggered the defense mechanisms. On the other hand, the flavonoids controlling the aroma and flavor are secondary metabolites, being synthesized in response to an excess of photoassimilation. The expression level of the transcripts related with flavonoid biosynthesis were increased in the “off” year leaves in relation to the “on” year ones in the olive tree. A similar gene expression pattern was observed with citrus buds, being based on flavonoids acting as a reservoir for the photoassimilation surplus. Taking account the higher expression of transcripts related to the oxidation-reduction, carbohydrate metabolism and mineral transport, together with flavonoid biosynthesis in “off” year leaves, we conclude that the nutritional status may be the principal key controlling the alternate bearing in the olive tree. Supporting the present work, a wide range of the genes targeted by the olive tree miRNA were found to be mainly involved in the carbohydrate metabolic pathways. Despite providing no selective advantage to the host, the plasmid is faithfully maintained at,60 copies per cell. The 2 mm plasmid achieves this by encoding copy-number amplification and partitioning systems, and by borrowing host cell machinery for its replication and segregation. Retention of the 2 mm plasmid at normal copy number is also dependent on the host cell process of sumoylation, the post-translational modification of target proteins with the small Orbifloxacin ubiquitin-related modifier protein SUMO. Sumoylation is an essential conserved eukaryotic function known to regulate diverse cellular processes by modulating the interactions, localization, or post-translational stability of substrate proteins. Like ubiquitin, SUMO must be activated in a series of enzyme-catalyzed steps Tulathromycin B before being conjugated to target proteins by the E2 conjugating enzyme Ubc9. Some target proteins require an E3 ligase for recruitment to Ubc9.
The TbMSP of bloodstream trypanosomes classified as belonging to the families are also found in the genomes
Importantly, 1,2fucosyltransferase transferase is present in A. deanei but not in the S. culicis dataset, and fucose residues were found in high amounts on glycoinositolphospholipid molecules of A. deanei, different from the observations for other trypanosomatids. Although the role of fucose is unknown, 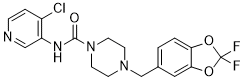 fucose and arabinose Albaspidin-AA transfer to lipophosphoglycan of Leishmania is noticed when the culture medium is supplemented with this carbohydrate, suggesting that fucose might have a specific role in A. deanei-insect interactions. Another glycosyltransferase found in both A. deanei and S. culicis genomes and involved in the N-glycosylation of asparagine residues is the dolichyl-diphosphooligosaccharide-protein glycosyltransferase, an oligosaccharyltransferase that is not classified in any of the above-mentioned families. The A. deanei and S. culicis DDOSTs contain the STT3 domain, a subunit required to establish the activity of the oligosaccharyl transferase complex of proteins, and they are orthologous to the human DDOST. These OTase complexes are responsible for transferring lipid-linked oligosaccharides to the asparagine side chain of the acceptor polypeptides in the endoplasmic reticulum, suggesting a conserved N-glycosylation among the trypanosomatids. Five different GalfT sequences are also present in the endosymbiont-bearing trypanosomatids, and all of them contain the proposed catalytic site, indicating genetic redundancy. Redundancy of GalfTs is commonly observed in many different trypanosomatid species, as different transferases are used for each linkage type. As b-galactofuranose has been shown to participate in trypanosome-host interactions, their presence in A. deanei and S. culicis might also indicate a role in the interaction with the insect host. However, no enzymes involved in synthesis of b-Galf-containing glycoconjugates are detected in our A. deanei dataset, despite reports of enzymes involved in b-Galf synthesis in Crithidia spp.. The presence of the N-terminal fatty acid acylation motif was found in some members of calpain-like cysteine peptidases, indicating that some of these peptidases are associated with membranes, as has also been shown for other members of the family. The relatively large amount of calpain-like peptidases may be related to the presence of the endosymbiont, which would require a more complex regulation of the cell cycle and intracellular organelle distribution, as cytosolic calpains were found to regulate cytoskeletal remodeling, signal transduction, and cell differentiation. A second large gene family in the A. deanei and S. culicis genomes encoding 3,4,5-Trimethoxyphenylacetic acid surface proteins with proteolytic activity is gp63. In our genomic analyses, we identified 37 and 9 genes containing sequences homologous to the gp63 of Leishmania and Trypanosoma spp. in the genomes of A. deanei and S. culicis, respectively. Proteins belonging to this group of zinc metalloproteases, also known as major surface protease or leishmanolysin, have been characterized in various species of Leishmania and Trypanosoma. Extensive studies on the role of this family in Leishmania indicate that they are involved in several aspects of host-parasite interaction including resistance to complement-mediated lysis, cell attachment, entry, and survival in macrophages.
fucose and arabinose Albaspidin-AA transfer to lipophosphoglycan of Leishmania is noticed when the culture medium is supplemented with this carbohydrate, suggesting that fucose might have a specific role in A. deanei-insect interactions. Another glycosyltransferase found in both A. deanei and S. culicis genomes and involved in the N-glycosylation of asparagine residues is the dolichyl-diphosphooligosaccharide-protein glycosyltransferase, an oligosaccharyltransferase that is not classified in any of the above-mentioned families. The A. deanei and S. culicis DDOSTs contain the STT3 domain, a subunit required to establish the activity of the oligosaccharyl transferase complex of proteins, and they are orthologous to the human DDOST. These OTase complexes are responsible for transferring lipid-linked oligosaccharides to the asparagine side chain of the acceptor polypeptides in the endoplasmic reticulum, suggesting a conserved N-glycosylation among the trypanosomatids. Five different GalfT sequences are also present in the endosymbiont-bearing trypanosomatids, and all of them contain the proposed catalytic site, indicating genetic redundancy. Redundancy of GalfTs is commonly observed in many different trypanosomatid species, as different transferases are used for each linkage type. As b-galactofuranose has been shown to participate in trypanosome-host interactions, their presence in A. deanei and S. culicis might also indicate a role in the interaction with the insect host. However, no enzymes involved in synthesis of b-Galf-containing glycoconjugates are detected in our A. deanei dataset, despite reports of enzymes involved in b-Galf synthesis in Crithidia spp.. The presence of the N-terminal fatty acid acylation motif was found in some members of calpain-like cysteine peptidases, indicating that some of these peptidases are associated with membranes, as has also been shown for other members of the family. The relatively large amount of calpain-like peptidases may be related to the presence of the endosymbiont, which would require a more complex regulation of the cell cycle and intracellular organelle distribution, as cytosolic calpains were found to regulate cytoskeletal remodeling, signal transduction, and cell differentiation. A second large gene family in the A. deanei and S. culicis genomes encoding 3,4,5-Trimethoxyphenylacetic acid surface proteins with proteolytic activity is gp63. In our genomic analyses, we identified 37 and 9 genes containing sequences homologous to the gp63 of Leishmania and Trypanosoma spp. in the genomes of A. deanei and S. culicis, respectively. Proteins belonging to this group of zinc metalloproteases, also known as major surface protease or leishmanolysin, have been characterized in various species of Leishmania and Trypanosoma. Extensive studies on the role of this family in Leishmania indicate that they are involved in several aspects of host-parasite interaction including resistance to complement-mediated lysis, cell attachment, entry, and survival in macrophages.
Imply that Purkinje cells initially inhibit OPC differentiation during their maturation by releasing Shh and then subsequently promote
Second, Shh stimulates OPC proliferation. Our results confirm the mitogenic effect of Shh on OPCs. These results also suggest that changes in Shh expression may control the timing of OPC differentiation. Thus, Purkinje cells, through Shh secretion, participate in the control of OPC proliferation in the cerebellum. The addition of vitronectin to immature Albaspidin-AA slices enhanced oligodendrocyte differentiation and vitronectin-blocking antibodies inhibited the effects of mature-slice induced differentiation on immature slices. Furthermore, when the slices cultures contained more Purkinje cells, levels of vitronectin were elevated. Only a slight difference between control and axotomized slices was detected likely because we are closed to the threshold of detection. Thus, our results clearly demonstrate that vitronectin is required for mature slices to promote oligodendrocyte differentiation in immature slices. Previous studies reported no effect of vitronectin on OPC differentiation. Moreover, it has been shown that the CG4 cell lines grown on vitronectin proliferate more that those grown on polylysine. However, combinations of vitronectin and mitogenic factors stimulate the differentiation of embryonic stem cells into oligodendrocytes, whereas vitronectin alone has no effect. Interestingly, Vitronectin inhibits the effects of Shh in other systems, through biochemical interactions in the spinal cord or the signaling pathway induced by its binding to integrin in cerebellar granule cells. Nevertheless, Shh and vitronectin may also control OPC differentiation through different mechanisms. Organotypic culture is an integrated system, in which cell interactions mimic those occuring in vivo, and is easier to manipulate than in vivo models. Furthermore only one type of neuron is myelinated in this system: the Purkinje cell. Using this system, we showed that the maturation of the Purkinje cell is involved in Tulathromycin B controlling the timing of oligodendrocyte differentiation. Indeed, our results suggest that Purkinje cells release different factors during their maturation, which have opposing effects on oligodendrocyte differentiation. This temporal regulation probably synchronizes the differentiation of oligodendrocytes and Purkinje cells. However, as discussed above, oligodendrocyte differentiation occurs even when the number of Purkinje cells is reduced. This suggests the presence of other differentiating factors in cultured slices or medium. Many of the factors known to affect oligodendrocyte development, such as TGF?, IGF-1, and progesterone, are present in the cerebellum. Oligodendrocyte differentiation 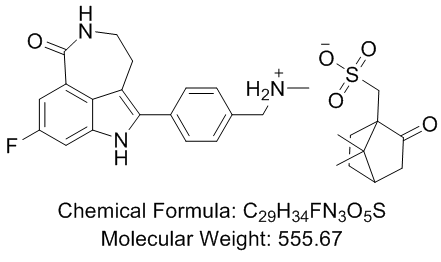 rates were lower in serum-free medium than in the presence of serum. Thyroid hormones are crucial for both oligodendrocyte and Purkinje cell differentiation. Among the factors involved in controlling OPC proliferation and differentiation, our study identifies two molecules with developmentally regulated expression patterns which are involved in the switch between OPC proliferation and oligodendrocyte differentiation. Vitronectin has been detected in active multiple sclerosis plaques and a decrease in Shh levels has been observed in the white matter of patients with multiple sclerosis. These actors are thus present, but it is unclear as to whether they are correctly synchronized. Determining how the timing of the various steps leading to myelination or remyelination is controlled is therefore still a major challenge.
rates were lower in serum-free medium than in the presence of serum. Thyroid hormones are crucial for both oligodendrocyte and Purkinje cell differentiation. Among the factors involved in controlling OPC proliferation and differentiation, our study identifies two molecules with developmentally regulated expression patterns which are involved in the switch between OPC proliferation and oligodendrocyte differentiation. Vitronectin has been detected in active multiple sclerosis plaques and a decrease in Shh levels has been observed in the white matter of patients with multiple sclerosis. These actors are thus present, but it is unclear as to whether they are correctly synchronized. Determining how the timing of the various steps leading to myelination or remyelination is controlled is therefore still a major challenge.