Unlike clonal cultures, these neurospheres were formed by aggregation, which results in culture heterogeneity.The minor transgene expression variability among Lomitapide Mesylate neurosphere cultures generated from littermate fetuses possibly occurred during the initial brain harvest. We did not specifically dissect the forebrain from each fetus. Nestin expressing neural SCs in the developing midbrain and hindbrain may have contributed to the transgene expressing cell population since CKIIa, the promoter driving tTA-transgenic human tau expression, is expressed throughout the brain at this developmental age. We saw more variability among independent experimental harvests than among cultures derived from littermate fetuses; we attribute this variation to inconsistencies inherent to IFA. Regardless, IFA consistently demonstrated that undifferentiated cells derived from rTg expressing fetal brains expressed human tau in a higher proportion of cells than those derived from rTg expressing fetal brains. Total brain homogenates Mechlorethamine hydrochloride indicate that rTg mice express comparable levels of transgenic tau as rTg. While the rTg cultures had a lower proportion of total cells expressing human tau, they contained a greater proportion of cells that expressed higher levels of tau. This feature of some transgenes is caused by position-effect variegation. Neurospheres, like the mice from which they were derived, may show this effect and could be useful models for screening transgene expression in founder lines. Consistent with the difference in transgene expression seen in undifferentiated neurospheres, differentiated cells derived from tauwt-expressing neurospheres expressed human tau in a higher proportion of cells than those derived from tauP301L-expressing neurospheres. Likely, non-transgene expressing progenitor cells gave rise to nontransgene expressing differentiated cells and transgene expressing progenitor cells differentiated into transgene expressing mature cells. Alternatively, tauP301L transgene expression may have decreased neural precursor survival. Since neurospheres proliferated and differentiated over several passages in both genotypes, and differentiated transgene expressing cell proportions mirrored that of the undifferentiated condition, evidence favors the former explanation. Some of the differences we saw between tauP301L and tauwt may stem from differences in transgene insertion sites. Neurosphere cultures from mouse lines expressing the same human tau variants, but at lower levels than rTg4510 and rTg21221, overlapped in the percentages of cells expressing human tau, but within the same range as our more extensively studies lines. It is likely that there are effects of transgene insertion site as well as the tau mutation. With the emergence of methodologies to culture neurospheres from human patients, experiments evaluating the culture system’s relevance and validity are crucial. Our data provide supporting evidence that neurospheres can reliably model phenotypes of their derived source over extended culture periods. The neurosphere culture system provides a robust assay for studying effects of external factors 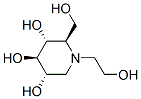 on the development and differentiation of the CNS, and the genetic susceptibility to neurological disorders. While it is unreasonable to expect that an in vitro system will fully recapitulate a complex disease process involving complex cell interactions, biologically relevant models that provide reproducible results are invaluable resources.
on the development and differentiation of the CNS, and the genetic susceptibility to neurological disorders. While it is unreasonable to expect that an in vitro system will fully recapitulate a complex disease process involving complex cell interactions, biologically relevant models that provide reproducible results are invaluable resources.
Category: clinically Small Molecule
Phenotypes of neurosphere cultures isolated from individual fetuses at different times reflected
An engineered tau variant that mimics constitutively phosphorylated tau by replacing Ser/Thr residues with glutamate, did not develop tau aggregates, neuronal loss, or behavioral abnormalities. In contrast, mice expressing tauR406W, a variant that remains hypophosphorylated compared to tauwt even in aged mice, developed age-dependent tau aggregates and memory and behavioral abnormalities. The hypophosphorylated tauP301L species in neurospheres and young mice may represent free tau not associated with microtubules. Tau bound to MTs acquires more phosphorylation than free tau as demonstrated by in vitro phosphorylation of tau in the Folinic acid calcium salt pentahydrate presence or Pimozide absence of MTs. Many of the MAPT exon 10 missense mutations that cause dementia, including P301L, reduce the ability of tau to interact with MT, and tauwt displaces mutant tau from MTs. The absence of tauP301L aggregates or neurofibrillary tangles in neurospheres and in young mice, despite phosphorylation at many of the sites most frequently phosphorylated in AD and FTD, also may correspond to unbound tau as MT association has been implicated as an important step for tau nucleation. The quantity of NFTs correlates with disease severity. However, recent studies have dissociated NFTs from neuronal death and decreased memory function, and instead suggest a deleterious effect of soluble tau. Phosphorylation of mouse tau also reflects appropriate phosphorylation corresponding to the differentiation state. Heavily phosphorylated 3R mouse tau is know to bind to nucleolar organizing regions in dividing cells and was also observed in neurospheres, indicating that human tau did not inhibit either the cellular machinery or kinases involved. This reinforces the same conclusion coming from the recapitulation of the genetic differences in human tau phosphorylation seen in neurosphere culture. We observed differences in filopodia-spine densities between tauwt and tauP301L differentiated cells. Developmentally, dendritic spine morphology evolves from long thin filopodia-spines to mature spines of various morphologies; during this transformation, filopodia-spine density decreases. We observed a slightly higher filopodia-spine density in Map2 and TUJ-1 double-positive cells from tauP301L cultures than those derived from tauwt or cultures that did not express MAPT. While the level of transgene expression within individual cells did not affect filopodia-spine density, we have not ruled out an effect of the transgene insertion site. Interestingly, in a separate P301L mouse line that that harbors the mutation in the longest mouse tau isoform, 4R2N, driven by the Thy1 promoter, young mice with “hypophosphorylated” tau have enhanced learning and memory and increased Long Term Potentiation in the dentate gyrus 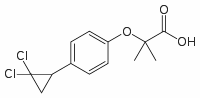 compared to controls. With age, the spine density of rTg4510 mice decreases and coincides with increased neuronal excitability. Whether or not the greater filopodiaspine density we observed in differentiated tauP301L cultures relates to enhanced LTP in young mice or neuronal vulnerability later in life in unknown, but warrants further exploration. We did not observe mislocalization of tauP301L to dendritic spines, as reported in aging mice or in transfected rat neuron cultures. We saw high levels of transgenic tau protein throughout neuritic projections in differentiated neurospheres from both human tau transgene genotypes. The presence of tau in the filopodia-spines of developing cells suggests that the phenomenon correlates with a dynamic morphology and does not necessarily indicate pathological injury.
compared to controls. With age, the spine density of rTg4510 mice decreases and coincides with increased neuronal excitability. Whether or not the greater filopodiaspine density we observed in differentiated tauP301L cultures relates to enhanced LTP in young mice or neuronal vulnerability later in life in unknown, but warrants further exploration. We did not observe mislocalization of tauP301L to dendritic spines, as reported in aging mice or in transfected rat neuron cultures. We saw high levels of transgenic tau protein throughout neuritic projections in differentiated neurospheres from both human tau transgene genotypes. The presence of tau in the filopodia-spines of developing cells suggests that the phenomenon correlates with a dynamic morphology and does not necessarily indicate pathological injury.
Contrast reveal that BiFC puncta localize at the plasma membrane in contact with the substrate
These puncta are essentially abolished by replacing the regulatory C-terminal tyrosine residue of Src by phenylalanine. In addition, mutations disrupting the SH3 binding motif in wild type PTP1B reveal a secondary site of interaction. Our results show for the first time physical interactions of ER-bound PTP1B with Src at puncta localized at the plasma Ginsenoside-F4 membrane in contact with the substrate, and further reveal that this interaction critically depends on the active site of PTP1B and the regulatory tyrosine 529 of Src. The results of the present paper illustrate a case of functional modulation in trans, among molecules located at the surface of the ER and plasma membrane, a phenomenon which may apply to a wide range of molecules and have impact in the regulation of several cell processes. In the present paper we used the BiFC technique to demonstrate direct physical interactions among ER-bound PTP1B and tyrosine kinases Src and Fyn at the plasma membrane. These interactions were revealed as bright fluorescence puncta associated with the ER. Our results strongly 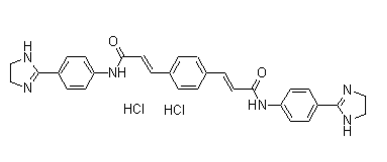 suggest that PTP1B, which is localized at the tip of dynamic ER tubules, was positioned close to the ventral membrane in contact with the substrate and interacted with Src at multiple puncta sites. We further show that mutations altering the active site of PTP1B and removing the negative regulatory residue of Src significantly reduced BiFC complex formation. These results suggest that ER-bound PTP1B releases Src from its negative regulation at random point contacts of the membrane/ substrate interface, leading to its activation and possibly recruitment to adhesion complexes. The subcellular localization of BiFC puncta is expected to be conditioned by the subcellular localization of each interaction partner. PTP1B is anchored to the cytosolic face of the ER membrane through a hydrophobic C-tail, allowing its mobility through the vast surface of the ER, and its interaction with substrates in the cytosol. In addition, dynamic changes of shape of the ER, dependent on its association with microtubules, extend the range of PTP1B interactions to a larger spatial scale, positioning PTP1B in the cell cortex, and therefore facilitating its encounter with substrates associated with the cytosolic face of the plasma membrane. Among these potential substrates, several laboratories including ours have identified the Src family of tyrosine kinases. Src kinases associate with plasma membrane by means of fatty acid modifications, and by protein protein interactions. In addition, a fraction of Src remains in the cytosol and another is associated with recycling endosomes. On this regard, the weak BiFC signal revealing the ER network, when using the wild type PTP1B, likely represents interactions with the freely diffusing pool of cytosolic Src. In contrast, bright BiFC puncta in the ER likely reflects interactions with spatially restricted Src and PTP1B molecules. We found that BiFC puncta did not co-localize with rab11, a recycling endosomes marker, and did not display directional movement in the cytoplasm, as expected for traffic carriers. Therefore, it is unlikely that BiFC puncta reflect interactions with an endosomal pool of Src. The fact that BiFC puncta were Cinoxacin retained in ventral membrane preparations, were visualized within the evanescent field produced by TIRF microscopy, and some colocalized with dark spots seen under SRIC microscopy, strongly suggest that they could represent spatially restricted interactions of ER-bound PTP1B with a subset of Src localized at the plasma membrane. Indeed, BiFC was significantly reduced when SrcTYN was used.
suggest that PTP1B, which is localized at the tip of dynamic ER tubules, was positioned close to the ventral membrane in contact with the substrate and interacted with Src at multiple puncta sites. We further show that mutations altering the active site of PTP1B and removing the negative regulatory residue of Src significantly reduced BiFC complex formation. These results suggest that ER-bound PTP1B releases Src from its negative regulation at random point contacts of the membrane/ substrate interface, leading to its activation and possibly recruitment to adhesion complexes. The subcellular localization of BiFC puncta is expected to be conditioned by the subcellular localization of each interaction partner. PTP1B is anchored to the cytosolic face of the ER membrane through a hydrophobic C-tail, allowing its mobility through the vast surface of the ER, and its interaction with substrates in the cytosol. In addition, dynamic changes of shape of the ER, dependent on its association with microtubules, extend the range of PTP1B interactions to a larger spatial scale, positioning PTP1B in the cell cortex, and therefore facilitating its encounter with substrates associated with the cytosolic face of the plasma membrane. Among these potential substrates, several laboratories including ours have identified the Src family of tyrosine kinases. Src kinases associate with plasma membrane by means of fatty acid modifications, and by protein protein interactions. In addition, a fraction of Src remains in the cytosol and another is associated with recycling endosomes. On this regard, the weak BiFC signal revealing the ER network, when using the wild type PTP1B, likely represents interactions with the freely diffusing pool of cytosolic Src. In contrast, bright BiFC puncta in the ER likely reflects interactions with spatially restricted Src and PTP1B molecules. We found that BiFC puncta did not co-localize with rab11, a recycling endosomes marker, and did not display directional movement in the cytoplasm, as expected for traffic carriers. Therefore, it is unlikely that BiFC puncta reflect interactions with an endosomal pool of Src. The fact that BiFC puncta were Cinoxacin retained in ventral membrane preparations, were visualized within the evanescent field produced by TIRF microscopy, and some colocalized with dark spots seen under SRIC microscopy, strongly suggest that they could represent spatially restricted interactions of ER-bound PTP1B with a subset of Src localized at the plasma membrane. Indeed, BiFC was significantly reduced when SrcTYN was used.
Revealed that membrane-bound wild type Src-GFP has lateral diffusion rates similar to lipid probes
Property impaired in active Src-Y527FGFP mutant, presumably due to interactions with other membrane proteins in a Src SH2-dependent manner. Interestingly, we found that BiFC puncta were significantly reduced when the Y529F mutant of Src was analyzed, suggesting that PTP1B targets plasma membrane Src on this residue. Our 3,4,5-Trimethoxyphenylacetic acid time-lapse experiments showed that BiFC puncta move apparently at random and locate at the tip of ER tubules, and under TIRFM they were frequently visualized as bright spots at one tip of comet-like Ginsenoside-Ro fluorescent structures, suggesting a “dipping down” of ER tubules towards the plasma membrane in contact with the substrate. Parallel analysis of GFPPTP1BDA showed similar results suggesting that PTP1B localization at the tip of ER tubules imposes a restriction to the spatial propagation of the interaction with Src molecules associated to the cytosolic side of the plasma membrane. A schematic view derived from our results is shown in Figure 6. ERbound PTP1B positioning in the cell cortex requires microtubules. The cometlike fluorescent figures observed under TIRFM suggest that ER tubules approach to the membrane at different angles, as previously shown for microtubules. In the context of the substrate trapping mutant PTP1BDA, interactions with plasma membrane-associated Src are stabilized, and therefore BiFC puncta enhanced. Mechanisms underlying the fusion and split of BiFC puncta, which were also seen for GFP-PTP1B, are currently unknown, but likely depend on the dynamics of ER tubules. Our analysis of BiFC was extended to YC-PTP1BWT/Fyn-YN and YC-PTP1BDA/Fyn-YN pairs. In both cases we found a positive BiFC signal that distributes in puncta, as it was described for Src. Fyn has a more tight association to the plasma membrane than Src due to additional palmitoylation, and quickly associates with the plasma membrane after being synthesized. This strengthens the view that BiFC puncta between PTP1B and Src kinases most frequently occur in association with the plasma membrane. However, we cannot completely rule out transient interactions between PTP1B and Src within the endosomal compartment. To elucidate this, a more extensive co-localization analysis with markers for different endosomes would be required, as well as high resolution double time-lapse analyses. In our study we showed that a truncation of the Src Nterminus, which removes the myristoylation target site and the polybasic motif involved in membrane association, eliminates the production of BiFC. Thus, membrane-bound Src is a requisite for BiFC to occur. Myristate is added to Src cotranslationally by the N-myristoyl-CoA-protein transferase enzyme. Beyond that, little is known about the regulation of Src myristoylation. We identified the active site of PTP1B and the Src tyrosine 529 at the C-tail as major determinants underlying BiFC. Tyrosine 529 is phosphorylated by the C-terminus Src kinase, Csk, and dephosphorylated by PTP1B and other tyrosine phosphatases. Replacement of tyrosine 529 by phenylalanine significantly reduced but did not completely eliminate BiFC puncta. In addition, a single mutation converting the PTP1B active site in a substrate trap significantly enhanced BiFC puncta throughout the cell, provided that the tyrosine 529 of Src remained unchanged. A second determinant contributing to BiFC is a proline-rich motif of PTP1B which fits the consensus sequence for class  II SH3 domain-binding motifs. Using PTP1BPA, a proline mutant in which the SH3binding motif was disrupted, moderately reduced the BiFC signal. Remarkably, PA mutation had no effect when combined with the substrate trap mutation DA.
II SH3 domain-binding motifs. Using PTP1BPA, a proline mutant in which the SH3binding motif was disrupted, moderately reduced the BiFC signal. Remarkably, PA mutation had no effect when combined with the substrate trap mutation DA.
The viral ORF sequences entity of the protein components of the tegument density has not been clearly determined
Moreover, the orientation and interaction of HCMV proteins in the virion have not been extensively studied. Much of what is currently known about the interactions among herpesvirus capsid and tegument proteins come from various studies involving protein assays and in particular, YTH analyses. Large-scale YTH analyses have also been applied to interactome studies of many organisms such as Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Saccharomyces cerevisiae, Plasmodium falciparum, andHelicobacter pylori. In complex organisms where their genome sizes are Albaspidin-AA relatively large, it is difficult to assess the importance of each individual protein to the systems. Therefore, it is important to map the global interactome to assess the true significance of each protein. Global genetic YTH analysis was also used to study the interactions between proteins encoded by vaccinia virus and five herpesviruses, which include herpes simplex virus 1, Varicella-zoster virus, EpsteinBarr virus, Gomisin-D murine cytomegalovirus, and Kaposi’s sarcoma-associated herpesvirus. Furthermore, the potential interactions among 5 capsid proteins and 28 tegument proteins of HCMV have recently been investigated using the YTH approach. These results have provided significant insights into the interactions among proteins encoded by herpesviruses. In this study, we have carried out a comprehensive YTH analysis to identify potential interactions among 56 HCMV virion proteins, which include 5 capsid proteins, 33 tegument proteins, and 18 envelope proteins. We have identified 79 pairs of potential interactions that are involved in viral capsid proteins, tegument proteins, and envelope proteins. Of the 79 interactions, 58 have not been previously identified to the best of our knowledge, while 21 of them have been reported. Forty-five of these 79 putative interactions were also identified in human cells by co-immunoprecipitation experiments. Our results indicate the presence of several HCMV proteins that serve as ”hubs” for interactions with numerous protein partners, thereby may function as an organizing center for connecting viral proteins in the mature virion and for recruiting other virion proteins. The interactions identified in this study provide a framework to study potential interactions between HCMV proteins and to investigate the functional roles of protein-protein interactions in HCMV virion assembly. We then used the locally written Unix-based scripts or automation of GCG package to analyze the obtained TowneBAC sequence and determine the coding sequences for ORFs that are 100 codons or longer. Each ORF was compared with the set of ORFs that had been predicted or found in all the HCMV strains for which sequences have been determined. This analysis suggested that the TowneBAC sequence encodes at least 150 ORFs with 100 amino acids or longer, and that all these ORFs align with those found in other HCMV strains. We initially selected an optimal PCR primer pair for each ORF. The primer pairs used for amplification of the viral sequences were constructed as follows. The forward primer contained the sequence immediately after the predicted translation initiation codon and 20�C25 additional nucleotides of coding sequence. The reverse primer contained the reverse complement of both the predicted translation termination codon and the preceding 20�C25 nucleotides at the end of the ORF. In addition, these primers also contain 15�C20 nucleotide common sequences that contain sites for restriction enzymes for cloning of the PCR products into the YTH screen vectors and the mammalian expression vectors. Each ORF encoding HCMV virion proteins was amplified individually by PCR. The amplified PCR products covered the entire ORFs minus the translation initiation codon, and were cloned into both the yeast two-hybrid screen ”prey” pGADT7 and ”bait” pGBKT7 vectors. We generated a collection of 118 constructs that 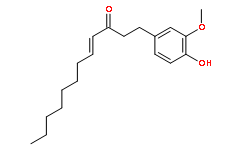 contained the sequences of the 56 HCMV ORFs, including those coding for exons 1 and 2 of UL89, the amino and carboxyl domains of UL48, and exons 1 and 2 of UL112.
contained the sequences of the 56 HCMV ORFs, including those coding for exons 1 and 2 of UL89, the amino and carboxyl domains of UL48, and exons 1 and 2 of UL112.